Exercise 1 – Develop an acquisition method
Task 5. Find optimum collision energy for MRM acquisition
Agilent 6400 Series Triple Quad LC/MS Familiarization Guide 31
2 Set up and run the worklist
(optional).
• Specify the data files as
iiiSulfamix MRM_xx.d, where
iii are your initials and xx is the
collision energy.
a Click the Worklist tab to make the
worklist visible.
b Add six samples to the worklist for
collision energies 10, 15, 20, 25, 30, 35.
c Mark the check box to the left of the
Sample Name for each of the three
samples.
d Click Worklist > Run.
• This step is optional because you
can use the six example data files
in the next step.
3 Compare the compound transition
intensities at different collision
energies.
• Open the MRM data files:
SulfamixMRM_10.d
SulfamixMRM_15.d
SulfamixMRM_20.d
SulfamixMRM_25.d
SulfamixMRM_30.d
SulfamixMRM_35.d
• Set the MRM chromatogram
extraction parameters as shown
at right for all transitions.
• Disable the TICs for clarity and
examine the peak intensities.
• Compare the intensities of each
compound transition obtained at
one collision energy with the
same compound transition
obtained at another collision
energy. (Do this in Overlaid
Mode with all the MRM
chromatograms.)
• Close the data files but don’t
save results.
• Refer to Table 4 on page 32 for
optimal method settings for
each compound.
a Open the Qualitative Analysis
program.
b Clear the Run ‘File Open’ actions...
check box.
c Open the MRM data files in the
Qualitative Analysis program.
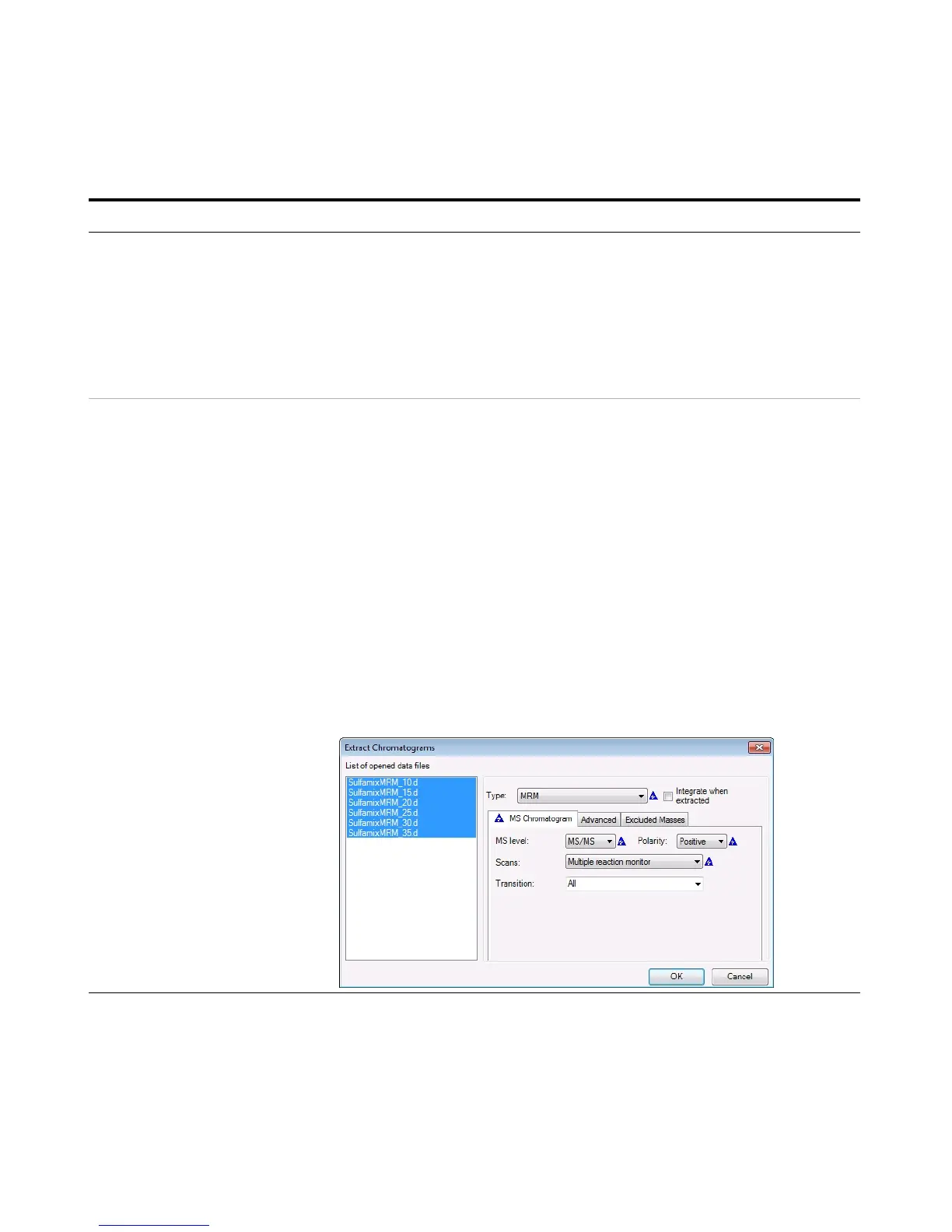
d Right-click the Chromatogram Results
window, and click Extract
Chromatograms from the shortcut
menu.
e To select all data files, click the last
file while holding down the Shift key.
f Enter the parameters as listed in the
example below, and click OK.
g Clear the TIC check boxes to make the
MRM chromatograms easier to view.
• Why a spectrum for MRM? It’s a
feature of the program to show
spectra even for MRM experiments
and can be quite handy for
comparing relative intensities of
product ions generated from the
same precursor.
• You can also click Chromatograms
> Extract Chromatograms to start
this dialog box.
Steps Detailed Instructions Comments

 Loading...
Loading...