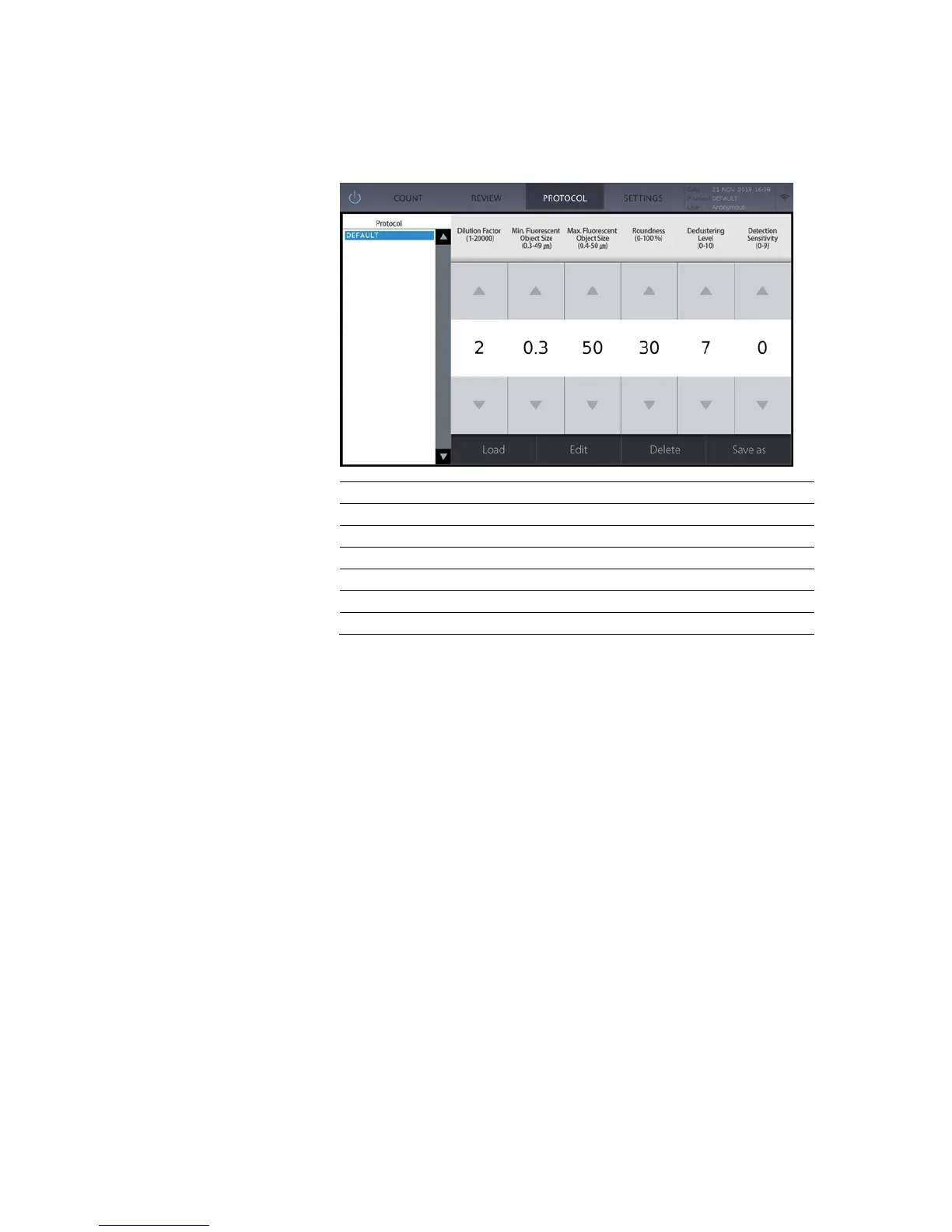
Counting parameters can be adjusted to customize protocols for specific bacteria.
Min Fluorescent Object Size (µm)
Max Fluorescent Object Size (µm)
*The DEFAULT protocol cannot be modified or deleted.
The dilution factor must be set properly prior to counting to ensure accuracy when
calculating the total cell concentration.
Final sample volume*
Dilution factor = ------------------------------------------ .
Bacterial suspension volume
*final sample volume = bacterial suspension volume + reagents volume
For users handling highly dense cultures, serial dilutions and several counts with
appropriately adjusted dilution factors will be necessary.
Fluorescent object size measures the diameter of the fluorescent signal from the nucleic
acid stains. This does not correspond to the physical size of the cell. This can be adjusted
in 0.1 µm increments between 0.3-1.0 µm and in 1 µm increments between 1-50 µm.
Set roundness according to the shape of sample. The counting algorithm will include
objects that are less round at lower values, which will include bacterial shapes such as
bacilli and vibrio. Higher values will include rounder shapes such as cocci.
The declustering function allows for the efficient detection of a cells that may grow in
clusters or chains. Higher values will lead to a higher sensitivity to clusters.
Detection sensitivity refers to the sensitivity of fluorescence detection. Higher values
detect fainter signals from weakly stained cells or smaller sample sizes but can also
increase noise.
 Loading...
Loading...