Appendix C - Calculations
C-3 BioMate 3 Operator’s Manual
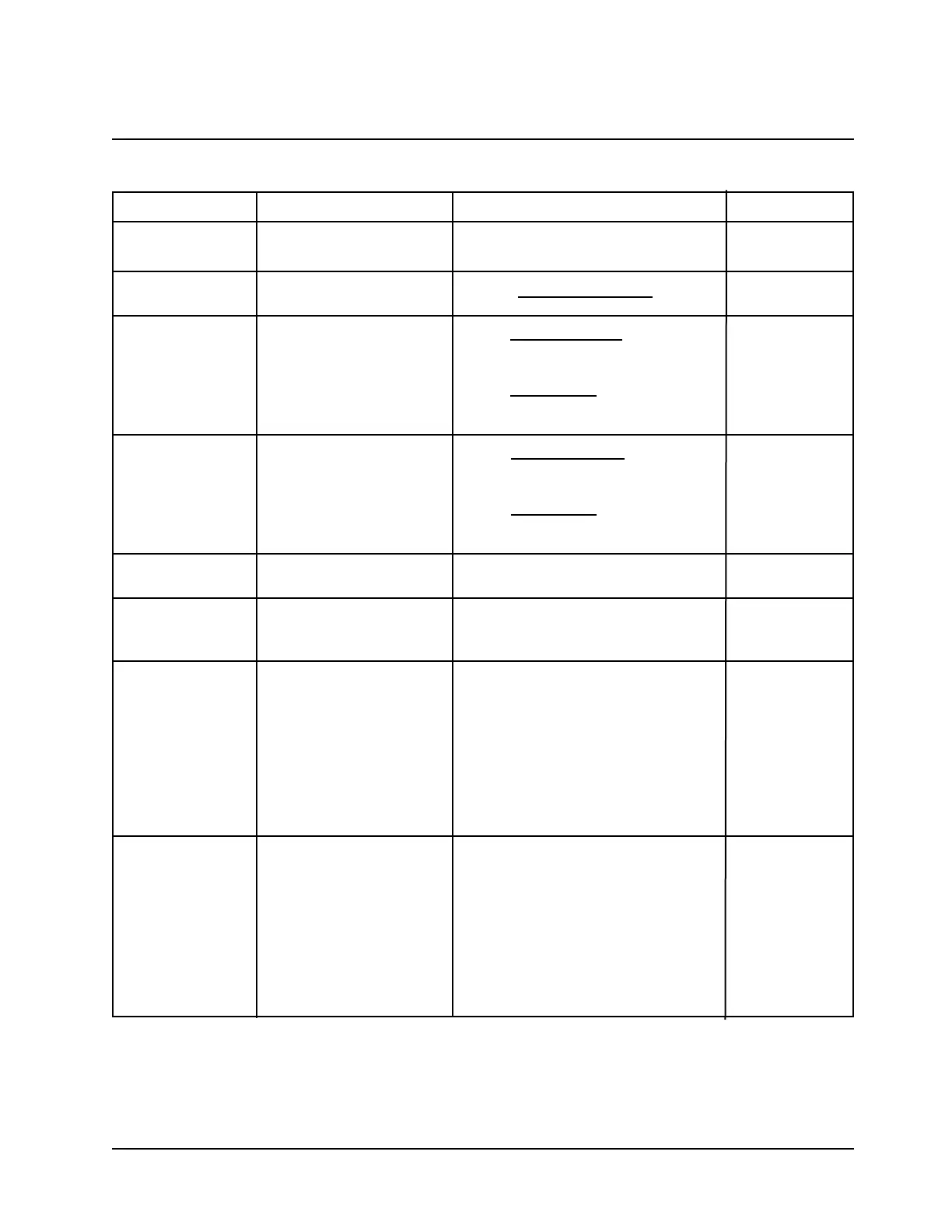
Calculation Entry Parameters Formula Displayed Units
# of bases Repetitive sequence of Count of total # of bases entered Length = # of
A, T (or U), G and C bases
%GC content Use AT (U) GC sequence %GC = # of (G + C) bases x 100 Percentage
entered above total # of AT(or U)GC
Molecular weight # units A, # units T, If entry does not include U: Molecular weight
# units G, # units C, MW = (312.2 x A) + (303.2 x T) = x daltons/M
# units U + (329.2 x G) + (289.2 x C) + 18.02
If entry does include U:
MW = (329.2 x A) + (306.2 x U)
+ (345.2 x G) + (305.2 x C) + 18.02
Absorptivity # units A, # units T, If entry does not include U: Extinction
ε (260) # units G, # units C, ε
260
= (15,200 x A) + (8,400 x T) coefficient =
# units U + (12,010 x G) + (7,050 x C) M
-1
cm
-1
If entry does include U:
ε
260
= (15,200 x A) + (9,900 x U)
+ (12,010 x G) + (7,050 x C)
Conversion Factor N/A Molecular Weight x 10
3
µg/mL
Extinction Coefficient
Calculation of T
m
: # units A, # units T, T
m
= 2(A + T) + 4(G + C) °C
Oligos up to # units G, # units C
20 bases in length
Calculation of Tm: • # units A, # units T, T
m
= 81.5C + 16.6log ((Na+)/ °C
DNA-DNA hybrids # units G, # units C (1+0.7(Na+)) + 0.41(%GC) –
• M = molarity of cation 500/L – P – 0.63(%formamide)
• Fraction GC = fraction
of G and C
• %form = %formamide
in the sample
• L = # of base pairs
• P = % mismatching
Calculation of Tm: • # units A, # units T, T
m
= 67°C + 16.6log ((Na+)/ °C
DNA-RNA hybrids # units G, # units C (1+0.7(Na+)) + 0.8(%GC) –
• M = molarity of cation 500/L – P – 0.5(%formamide)
• Fraction GC = fraction
of G and C
• %form = %formamide
in the sample
• L = # of base pairs
• P = % mismatching
Calculations
Table 2 Calculations for BioMate Oligo Calculator
 Loading...
Loading...