Chapter 5 Analyze the Relative Standard Curve Experiment
View the QC Summary
Applied Biosystems 7500/7500 Fast Real-Time PCR System Getting Started Guide for Relative Standard Curve
and Comparative C
T
Experiments
104
Notes
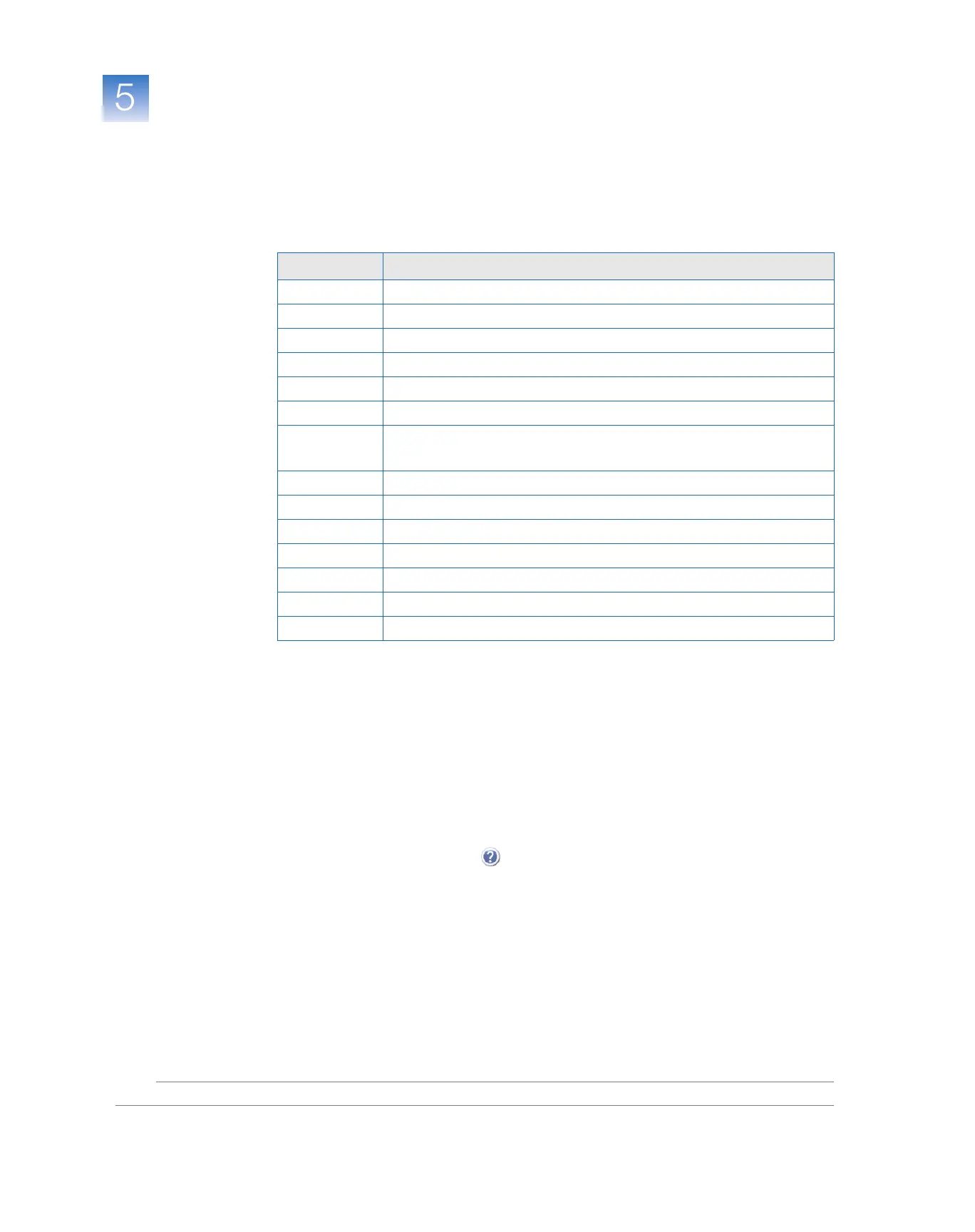
Possible Flags
For standard curve experiments, the flags listed below can be generated by the
experiment data.
If a flag does not appear in the experiment, its frequency is 0. If the frequency is >0, the
flag appears somewhere in the experiment; the well position is listed in the Wells column.
Analysis
Guidelines
When you analyze your own standard curve experiment:
• In the Flag Details table, click each flag that has a frequency >0 to display details
about the flag. If needed, click the troubleshooting link to view information on
correcting the flag.
• You can change the flag settings:
– Adjust the sensitivity so that more wells or fewer wells are flagged.
– Change the flags that are applied by the 7500 software.
For More
Information
For more information on the QC Summary screen or on flag settings, open the
7500 Software Help by clicking or pressing F1.
Flag Description
AMPNC Amplification in negative control
BADROX Bad passive reference signal
BLFAIL Baseline algorithm failed
CTFAIL C
T
algorithm failed
EXPFAIL Exponential algorithm failed
HIGHSD High standard deviation in replicate group
MTP Multiple Tm peaks
Note: This flag is displayed only if the experiment includes a melt curve.
NOAMP No amplification
NOISE Noise higher than others in plate
NOSIGNAL No signal in well
OFFSCALE Fluorescence is offscale
OUTLIERRG Outlier in replicate group
SPIKE Noise spikes
THOLDFAIL Thresholding algorithm failed
 Loading...
Loading...