DRAFT
September 1, 2004 11:39 am, CH_End-Point.fm
Chapter 5 Analyzing End-Point Data
5-6 Applied Biosystems 7900HT Fast Real-Time PCR System and SDS Enterprise Database User Guide
Overview
Allelic
Discrimination
on the 7900HT
Instrument
The Applied Biosystems 7900HT Fast Real-Time PCR System supports allelic
discrimination using TaqMan
®
probes. Allelic discrimination is the process by which
two variants of a single nucleic acid sequence are detected in a prepared sample.
Allelic discrimination chemistry can be used for single-nucleotide polymorphism
(SNP) detection.
Employing the
5´ Nuclease
Assay for Allelic
Discrimination
Allelic discrimination on the 7900HT instrument is made possible through the use of
the fluorogenic 5´ nuclease assay (see page D-2). During the PCR, the fluorogenic
probes anneal specifically to complementary sequences between the forward and
reverse primer sites on the template DNA. Then during extension, AmpliTaq Gold
®
DNA polymerase cleaves the probes hybridized to the matching allele sequence(s)
present in each sample. The cleavage of each matched probe separates the reporter dye
from the quencher dye, which results in increased fluorescence by the reporter. After
thermal cycling, the plate is run on the 7900HT instrument, which reads the
fluorescence generated during the PCR amplification. By quantifying and comparing
the fluorescent signals using the SDS software, it is possible to determine the allelic
content of each sample on the plate.
Mismatches between a probe and target reduce the efficiency of probe hybridization.
Furthermore, AmpliTaq Gold
®
DNA polymerase is more likely to displace the
mismatched probe than to cleave it, releasing the reporter dye. By running the extension
phase of the PCR at the optimal annealing temperature for the probes, the lower melting
temperatures (T
m
) for mismatched probes minimizes their cleavage and consequently
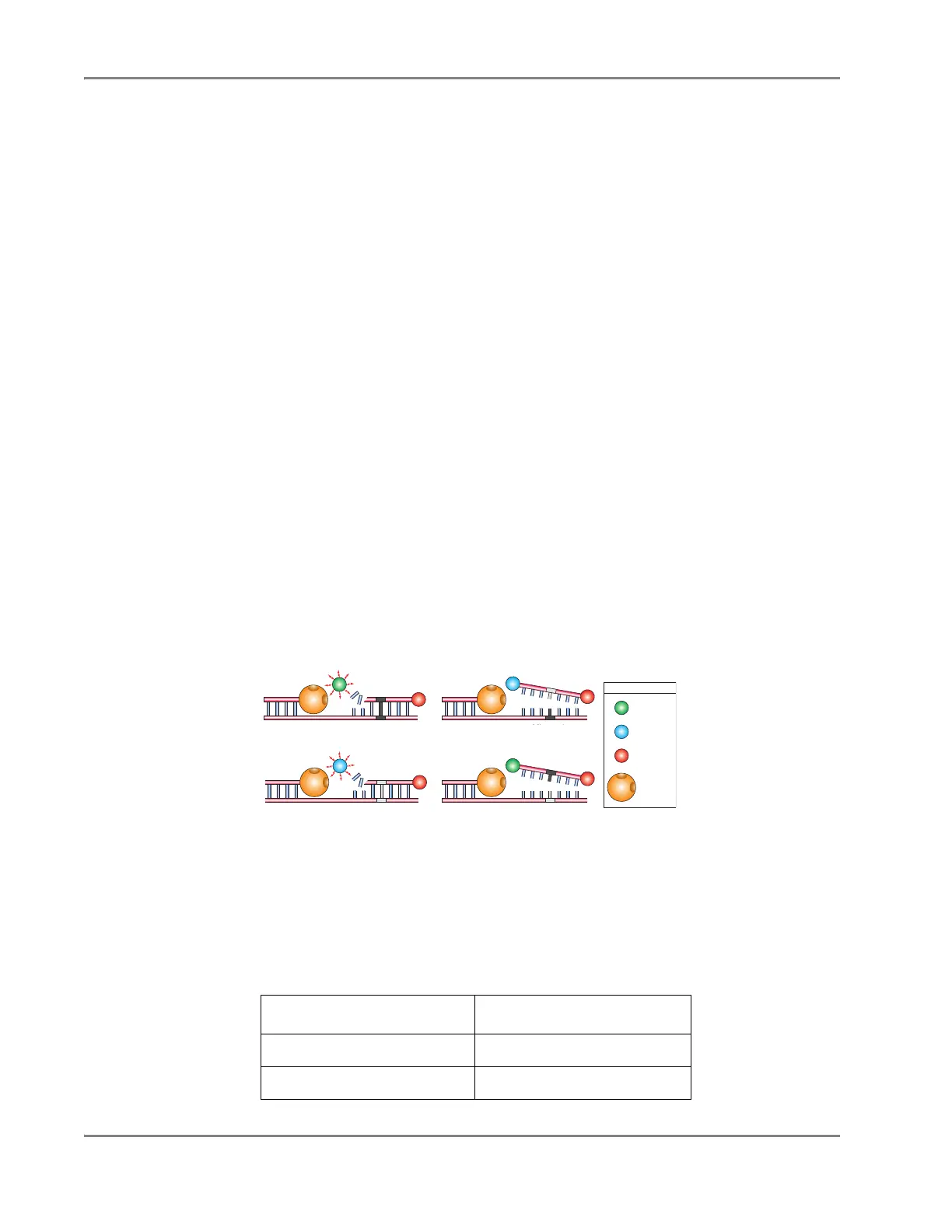
their fluorescent contribution. Figure 5-3 illustrates results matches and mismatches
between target and probe sequences in TaqMan
®
PDARs for AD assays (Livak
et al.
, 1995; Livak
et al.
, 1999).
Figure 5-3 Target and Probe Sequence Interaction
Table 5-1 shows the correlation between fluorescence signals and sequences present
in the sample.
Table 5-1 Signal and Sequence Correlation
A substantial increase in… Indicates…
VIC
®
dye fluorescence only homozygosity for Allele X.
FAM
™
dye fluorescence only homozygosity for Allele Y.
Allele
X
Match
V
Q
Mismatch
Mismatch
Q
V
F
Q
Allele
Y
Match
Q
F
Legend
AmpliTaq
Gold DNA
Polymerase
VIC
FAM
Quencher
Q
F
V
GR1556
Probe-target sequence
Probe-target sequence
mismatch (higher T
m
)
Probe-target sequence
Probe-target sequence
mismatch (higher T
m
)
 Loading...
Loading...