Proton Driven Spin Diffusion (PDSD)
User Manual Version 002 BRUKER BIOSPIN 149 (327)
Processing Parameters 11.3.1
Process with xfb.
Adjust the Rotational Resonance Condition for DARR/RAD 11.4
1. Load the Adamantane sample, spin at the same speed as desired for your
sample, match and tune, use a suitable cp setup (same as in section
11.3).
2. Set CPDPRG2 to cw.
3. Use the au program calcpowlev to calculate the power level required for a pro-
ton decoupling RF field of n × masr, using p3 and pl12 as reference values.
Refer to chapter
"Basic Setup Procedures" on page 55 for more informa-
tion).
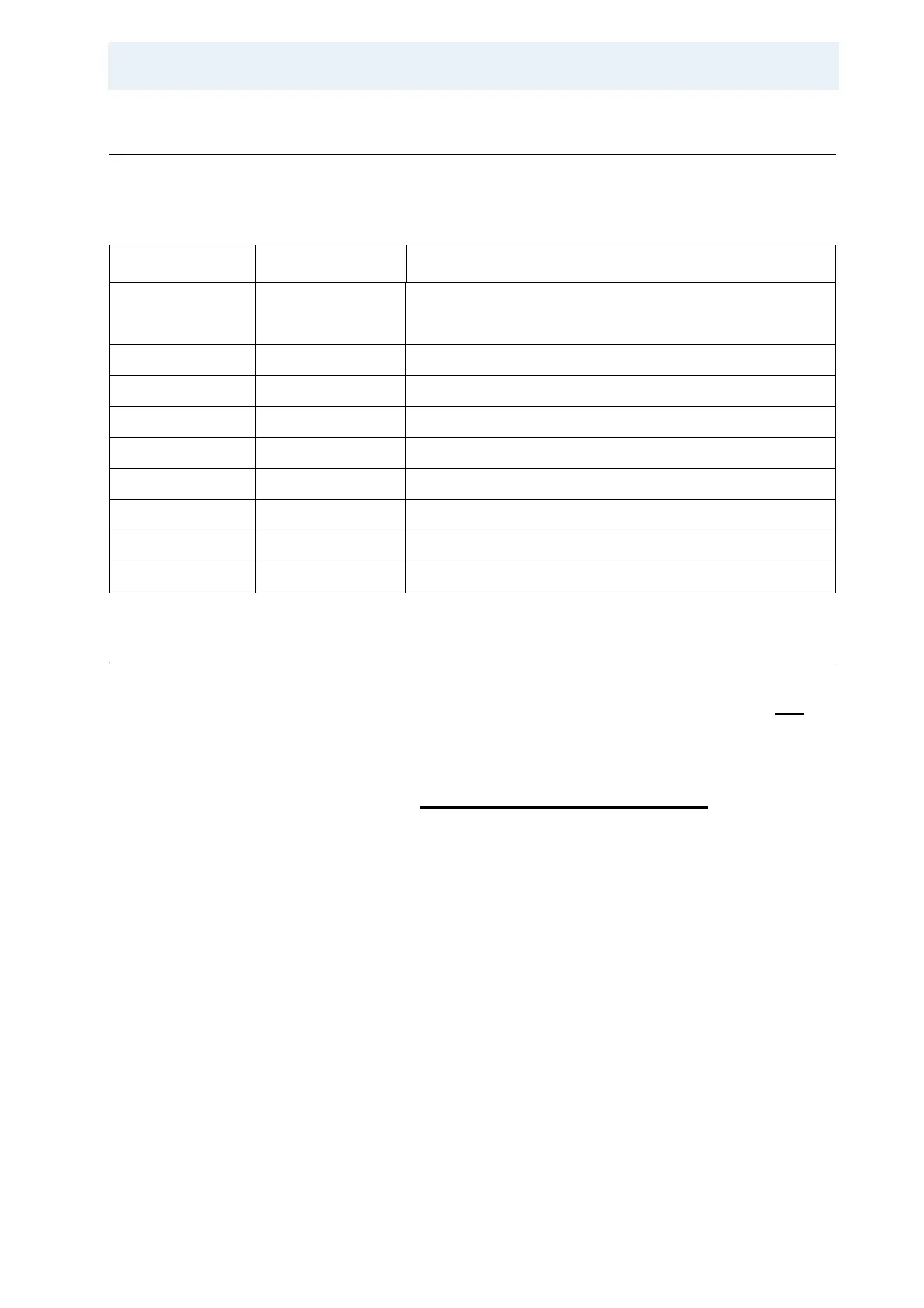
Table 11.2. Processing Parameters
Parameter Value Comment
F2 acquisition
13
C ************** Left column.
SI 1k Number of complex points in direct dimension.
WDW QSINE Apodization in t2.
SSB 2-3
PH_mod pk
F1 indirect
13
C ************** Right column.
SI 512 Number of complex points in indirect dimension.
WDW QSINE Apodization in t1.
SSB 2-3
PH_mod pk
 Loading...
Loading...