Setting
Description
Quality Threshold
• Basecall Assignment (ambiguous bases):
– Do not assign Ns to basecalls
– Assign Ns to basecalls with QV<15—Bases with a QV less than the
threshold display N instead of the base letter
• Ending base—Last base on which to perform basecalling:
• At PCR Stop
• After X number of Bases
• After X number of Ns in X number of Bases
• After X number of Ns
Note: If you have PCR products with sequences that end while data is still
being collected, select the At PCR Stop check box.
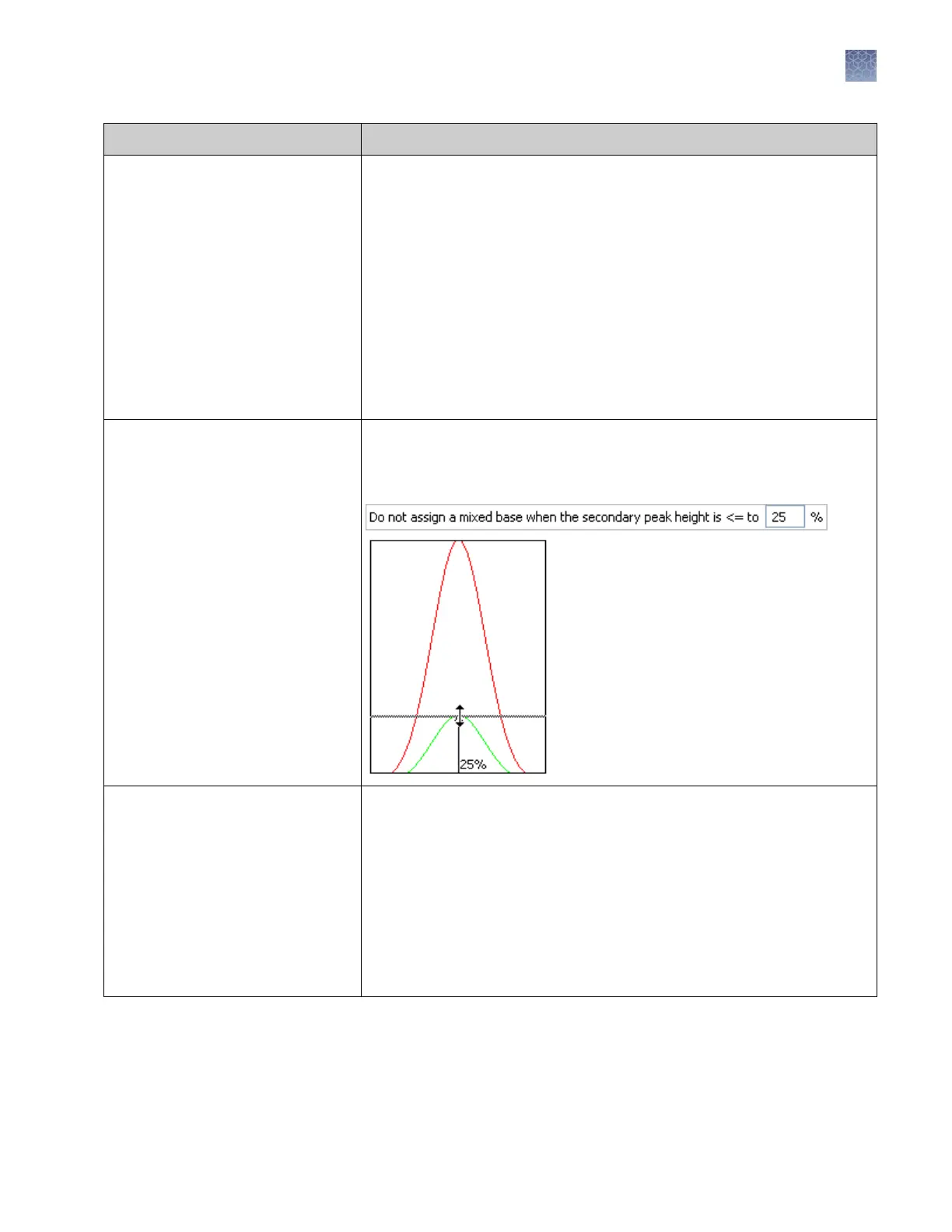
Mixed bases threshold When enabled, determines the secondary peak height ratio where the
secondary peak is considered a potential mixed base. Reaching the threshold
is a necessary but not sufficient condition for the basecalling algorithm to
call a mixed base.
Analyzed Data Scaling Determines scaling of the processed traces. This parameter does not affect
the ac
curacy of the basecalling.
• True Profile—The processed traces are scaled uniformly so that the
average height of peaks in the region of strongest signal is about equal
to a fixed value. The profile of the processed traces will be very similar to
that of the raw traces.
• Flat Profile—The processed traces are scaled semi-locally so that the
average height of peaks in any region is about equal to a fixed value. The
profile of the processed traces will be flat on an intermediate scale
(> about 40 bases).
Chapter 7 Manage library resources
Basecalling protocols library (primary analysis—sequencing)
7
3500/3500xL Genetic Analyzer User Guide—Data Collection Software v3.1
177
 Loading...
Loading...