[2]
T
he maximum number of contiguous bases in the analyzed sequence with an average QV ³20, calculated over a sliding window 20 base pairs
wide from an AB Long Read Standard sequencing sample. This calculation starts with base number 1. The read length is counted from the
middle base of the 1st good window to the middle base of the last good window, where a “good” window is one in which the average QV ³20.
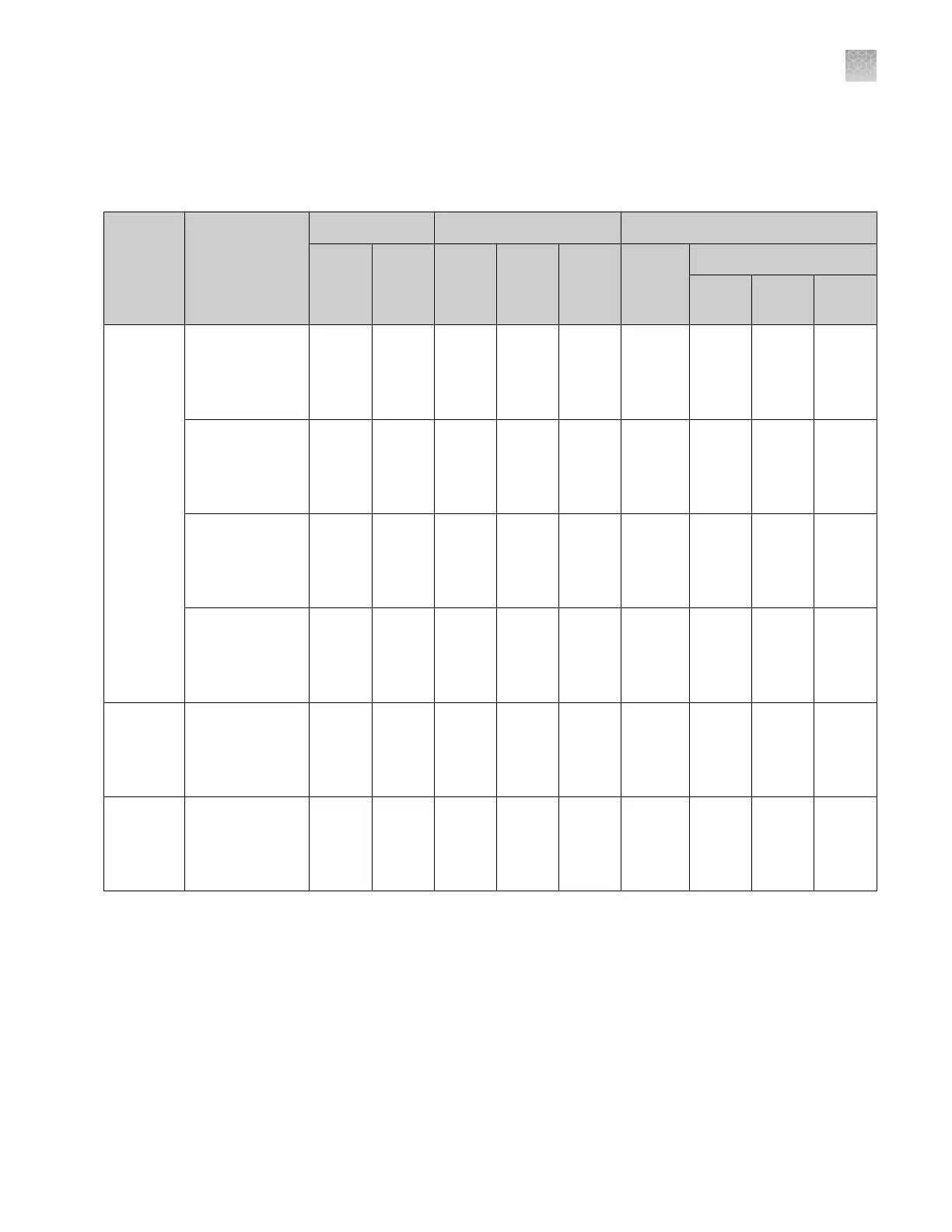
Table 6 Fragment and HID analysis run modules
Run
modul
e
type
Run module
name
Configuration 23 hours throughput
[1]
Performance
Cap.
length
(cm)
Pol.
type
Run
time
(min)
3500
(8-cap.)
3500xL
(24-
cap.)
Range
[2]
Sizing Precision
[3]
50bp-
400bp
401bp-
600bp
601bp-
1200bp
Frag.
analysis
FragmentAnalysi
s50_POP7
FragmentAnalysi
s50_POP7xl
50 POP-7
™
£40 ³280 ³840 £40
to ³520
<0.15 <0.30 NA
[4]
FragmentAnalysi
s50_POP6
FragmentAnalysi
s50_POP6xl
50 POP-6
™
£100 ³112 ³336 £20
to ³550
<0.15 <0.30 NA
[4]
FragmentAnalysi
s36_POP4
FragmentAnalysi
s36_POP4xl
36 POP-4
™
£35 ³312 ³936 £60
to ³400
<0.15 NA
[4]
NA
[4]
FragmentAnalysi
s36_POP7
FragmentAnalysi
s36_POP7xl
36 POP-7
™
£30 ³368 ³1104 £60
to ³400
<0.15 NA
[4]
NA
[4]
Frag.
analysis
FragAnalysis36_
POP6
FragAnalysis36_
POP6xl
36 POP-6
™
£60 ³184 ³552 £60
to ³400
<0.15 <0.30 NA
[4]
Long frag.
analysis
LongFragAnalysi
s50_POP7
LongFragAnalysi
s50_POP7xl
50 POP-7
™
£125 ³88 ³264 £40
to ³700
<0.15 <0.30 <0.45
Appendix B Run modules and dye sets
Run modules
B
3500/3500xL Genetic Analyzer User Guide—Data Collection Software v3.1
283
 Loading...
Loading...